- Overview
- AI Segmentation
- Features
- Speed
- Additional Modules
- Required Hardware
- Security
Overview
 sliceOmatic, is an easy-to-use, powerful and affordable medical image analysis software
that enhances researchers' ability to measure, segment and analyze
multi-slice scanner data.
sliceOmatic, is an easy-to-use, powerful and affordable medical image analysis software
that enhances researchers' ability to measure, segment and analyze
multi-slice scanner data.
"SliceOmatic is a unique software package that makes the segmentation process easy, intuitive, and highly interactive.
I can't imagine using anything else."
B.J. Fregly, Computational Biomechanics Lab, University of Florida
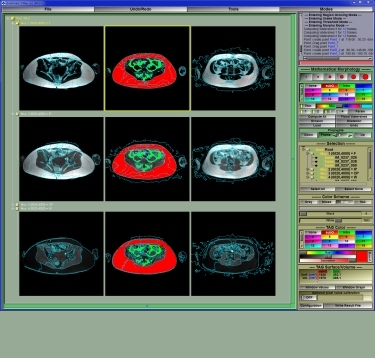
SliceOmatic offers you the best segmentation process to date. Previously segmentation with MRI (Magnetic Resonance) modality was a highly labor intensive task. With sliceOmatic, morphological operators can effortlessly perform tissue segmentation. Furthermore, studies have proven that this technique is generating consistent results.
NEW: AI with Python - The AI deep learning modules for segmenting any slices in any modalities.
The new AI with Python modules enable you to train your own AI with your own images. You can work with individual slices or with 3D volumes. The graphic interface make the training easy and intuitive for any AI novice, while also enabling power users to customize the python code used for the AI models and their metrics. Once you trained your AI, you can use it to segment images in as little as 20 msec. per slice with an Nvidia card, (about 10 times slower without).
But don't take our word for it, download sliceOmatic from this web site and try it out for yourself! Without a license the program does have a few limitations (it will not let you save any results) but you can take it for a spin and see how easy it is to segment your data with it.
It is used by satisfied customers around the world:
"Ah, SliceOmatic is a work of art! It lets me take my less-than-perfect MRI data and make
something beautiful out of it. I love the way it really lets you take a very close look at your data.
The screen space is used very efficiently, the tools are kept out of the way and the data is front
and center. The ability to quickly threshold and tag your data is great. It is then very easy to flip
between morpho and edit mode and finish off the tagging. The 3d rendering and .stl file writing
complete the package. The volume calculations are right on the money. While sliceO hasn't
saved my life, it sure has helped save my job!
I recommend it to everyone who wants to tag MRI data..... "
David Maltbie, Procter & Gamble.
"We have been delighted with our Tomovision programs. The Slice-O-Matic program has far exceeded
our expectations in terms of ease of use and quality of the output. We have found many more uses
for this program than we ever expected. We have encouraged other research institutions to consider
using Slice-O-Matic in their research. Thanks for the great product and the great service."
Judy Weltman, General Clinical Research Center, University of Virginia.
AI Segmentation
The AI with Python modules:|
These modules are included with sliceOmatic. The AI modules will let you analyze slices one by one or as 3D volumes.
|
|
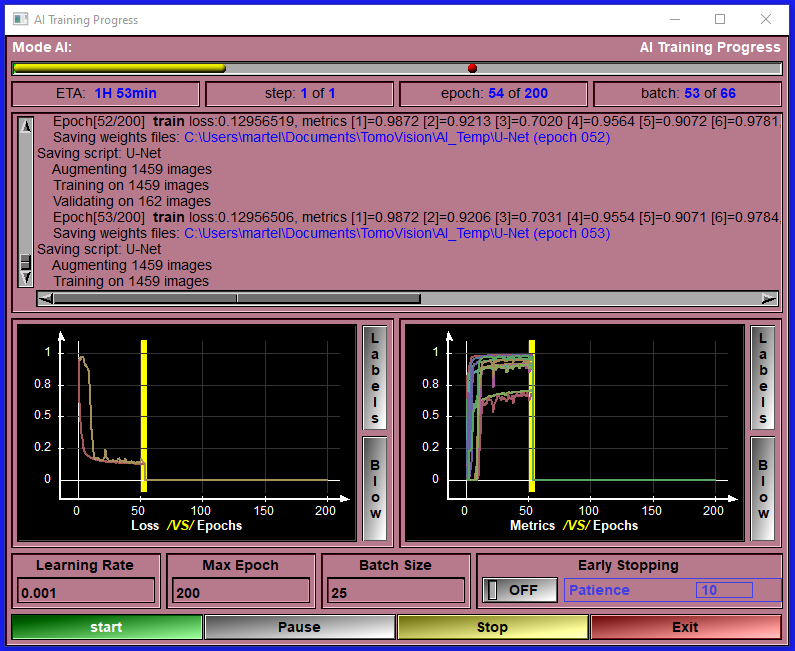
The AI modules enable you to train your own AI with your data. The AI modules can be used to segment images with pre-defined trained AI weights. If none of the available pre-trained weights match your requirements, you can train your own AI with your own data directly in these module. Everything you need to train an AI is provided. Just follow a few simple configuration steps, start the training, wait for it to finish (that step may need some patience...) save the resulting weights and use the newly trained AI to segment your images. You can use this process iteratively. First do some images manually or semi-automatically with the standard sliceO's interface, then train an AI with these few images. Use the trained AI as a first step in your segmentation process, correcting any errors with the sliceO many segmentation modules. Then when you have more images, re-train the AI. Eventually your AI will be good enough to do the segmentation of your images automatically. If you are familiar with AI technologies and can code in Python, these modules let you costumize the AI models and metrics as you please. |
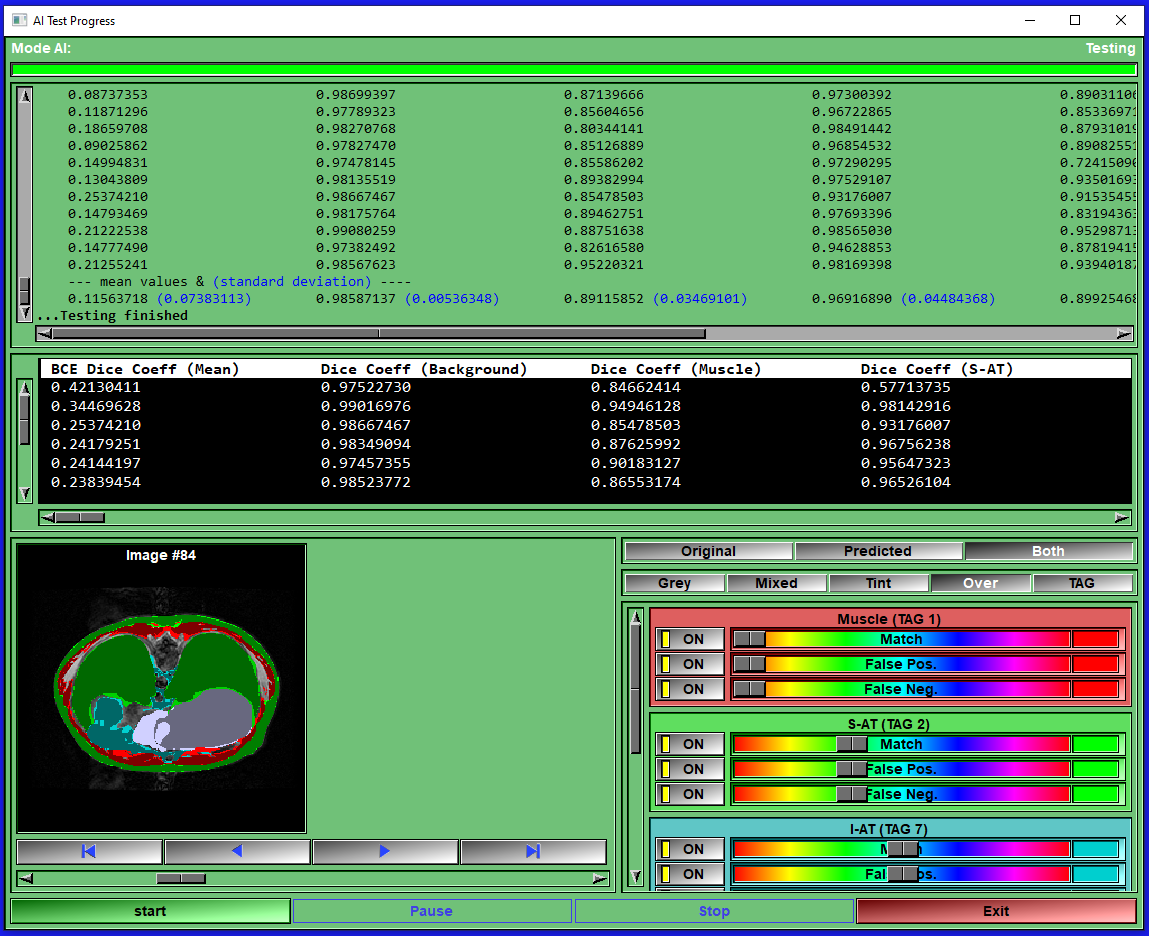
|
The ABACS module:
|
At the request of the makers of ABACS, the module is no longer available.
|
Features
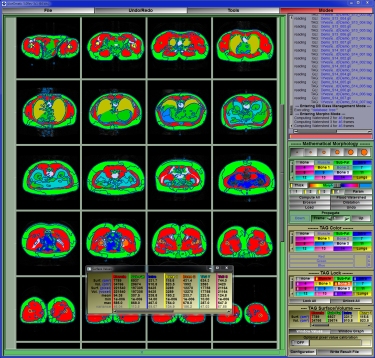
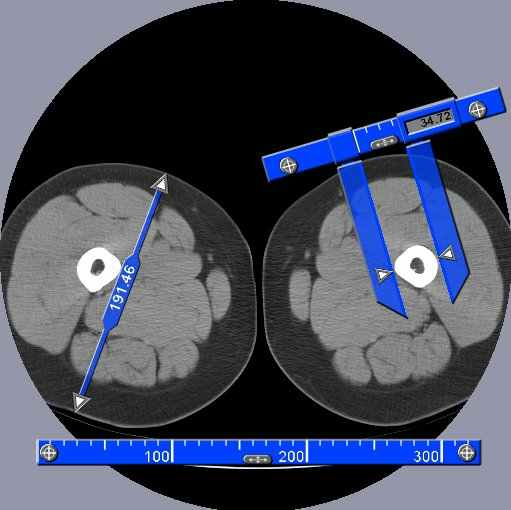
The usual tools: SliceOmatic has, of course, all the usual tools that any good image analisys program should offer. These include tools to view the images in any direction in multiple windows, tools to filter the images, tools to measure distances, angles and surfaces on the images, and a lot more of the "basic" stuff.SliceOmatic's Segmentation:
The process of identifying each tissue inside an image is known as "segmentation". And this is where sliceOmatic outshines the competition: its segmentation tools.
sliceOmatic balances powerful segmentation tools with a highly interactive interface, enabling you to completely control the segmentation process.
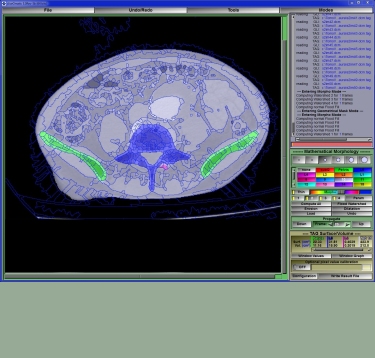
Different modalities or even different tissues within the same slice are best segmented with different techniques. SliceOmatic makes it easy for you to use the appropriate technique for each specific task. For example, in an MRI slice of the abdomen, you may want to use mathematical morphology to segment the sub-cutaneous fat and region-growing to segment the intra abdominal fat.
sliceOmatic is a research tool:
As a researcher, analysing the images is only a means to an end. What you really want is getting accurate anatomical volumes. SliceOmatic is designed as a research tool. In addition to helping you create the great images for these "wow" slides in your presentations, it will export the surfaces and volumes of each segmented tissue in a spread-sheet compatible file.
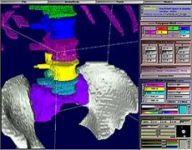
sliceOmatic and Biomechanics:
Maybe your area of research is biomechanics? Then you will want to use sliceOmatic to create polygonal meshs and/or countour lines from the segmented data. These can then be exported in a number of standard formats to be used either in finite element analisys or to create stereo-lithographic models.
Flexibility:
The wide range of formats that sliceOmatic supports makes it an ideal tool to both view and analyze all your images. You can see the list of supported image formats .
"I wanted to report back to you the outstanding results obtained with
the new slice0matic software. The system works beautifully and the new
software greatly eases analysis of MRI scans. Please extend my thanks to
Yves for a job well done."
Dr. Steve Heymsfield - St Luke Roosevelt Hospital, NY.
Speed
AI segmentation:The AI segmentation modules let you analyze slices in as little as 20msec per slice!
Manual segmentation:
In a technical publication, Dr Bonekamp and his colleagues compared sliceOmatic with 4 other image analysis packages. Not only did they found that sliceOmatic was among the easiest to use and had the best graphical user interface, but they also found that segmenting adipose and visceral fat with sliceOmatic was at least 3 time faster than with any of the other software.
| sliceOmatic | NIHImage | Analyze | Hippofat | EasyVision |
| 32 | 322.5 | 100 | 120 | 97.5 |
* From: S Bonekamp, P Ghosh, S Crawford et all, Quantitative comparison and evaluation of software packages for assessment of abdominal adipose tissue distribution by magnetic resonance imaging , Int J of Obesity, 32: pp.104, 2008
"[...] We consider sliceOmatic a very good choice for segmentation of abdominal
adipose tissue on MR images. [...]"
S Bonekamp, P Ghosh, S Crawford et all,
Quantitative comparison and evaluation of software packages for assessment of abdominal
adipose tissue distribution by magnetic resonance imaging
,
Int J of Obesity, 32:100-111, 2008
Additional Modules
The default installation of SliceOmatic already come with a lot of modules, but since we did not wanted to clutter the menus with things you may not need, a lot more are not selected to install by default.They are present in the installation package, juts not selected.
If you think you may have use of them, just select them at installation.
- Tool: Automation
- Tool: CT Gradient
- Tool: DICOM Compare
- Tool: DICOM Info
- Tool: Pixel Interpolation
- Class: Border
- Class: Clone
- Class: Gradient Amplification
- Class: RR2D
- Class: RR3D
- Mode: Histogram Segmentation
- Mode: TAG Interpolation
- Mode: MR Spectroscopy
Tool: Automation
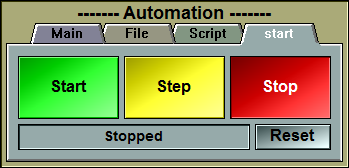 This tool enable you to automate some actions. It can be used to read files and perform scripts for each files, or perform scripts for each frames,
or simply perform scripts a number of times.
For example it can be used to take one snapshot of each frames, or to create an animation of a 3D object rotating, or in an industrial setting,
automate the analysis of images acquire from a continuous process.
This tool enable you to automate some actions. It can be used to read files and perform scripts for each files, or perform scripts for each frames,
or simply perform scripts a number of times.
For example it can be used to take one snapshot of each frames, or to create an animation of a 3D object rotating, or in an industrial setting,
automate the analysis of images acquire from a continuous process.
For more information, please consult the sliceOmatic 5.0 Online User's Manual
Tool: CT Gradient

 This tool is used to limit the gradient computation to a threshold range.
This tool is used to limit the gradient computation to a threshold range.
Some of the segmentation modes (Morpho Á Snakes) are based on the gradient of the pixel values. For CT images, we can also limit the gradient computation to a desired threshold, this give us a better segmentation.
For more information, please consult the sliceOmatic 5.0 Online User's Manual
Tool: DICOM Compare
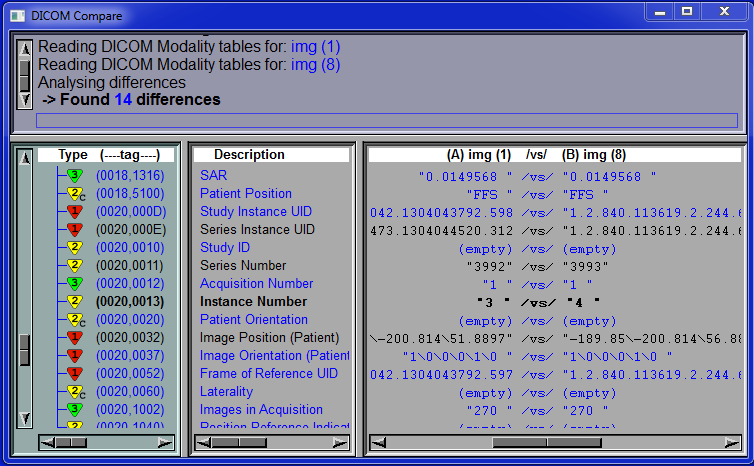 This tool integrate the standalone tool DICOM_Compare
from TomoVision inside sliceOmatic. It enable you to see the DICOM tags associated with any of the loaded frames.
You can either compare these tags with the DICOM definition for the same modality or with the tags of another frame.
This is very useful if you want to use the Mixer, Multiplex or DICOM Tree classes.
This tool integrate the standalone tool DICOM_Compare
from TomoVision inside sliceOmatic. It enable you to see the DICOM tags associated with any of the loaded frames.
You can either compare these tags with the DICOM definition for the same modality or with the tags of another frame.
This is very useful if you want to use the Mixer, Multiplex or DICOM Tree classes.
For more information, please consult the sliceOmatic 5.0 Online User's Manual
Tool: DICOM Info
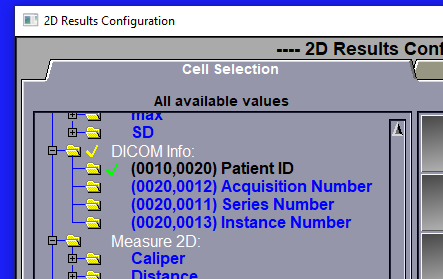 This tool is designed to work in conjuction with the "2D Results" tool.
It enable to save up to 4 DICOM tag values in the Excel result file.
This tool is designed to work in conjuction with the "2D Results" tool.
It enable to save up to 4 DICOM tag values in the Excel result file.
The 4 tags that are defined in the interface of the "DICOM Info" tool will be seen by the configuration page of the "2D Results" tool and can be selected. If some of the 4 tags are selected for the result file, the value of these DICOM tags, for each selected slice, will be saved in the Excel result file.
For more information, please consult the sliceOmatic 5.0 Online User's Manual
Tool: Pixel Interpolation


This module was developed by TomoVision for Yajima Nobuhiko of the Image Labo in Japan.
This tool enable you to "smooth" the images at higher magnifications.
For more information, please consult the sliceOmatic 5.0 Online User's Manual
Class: Border
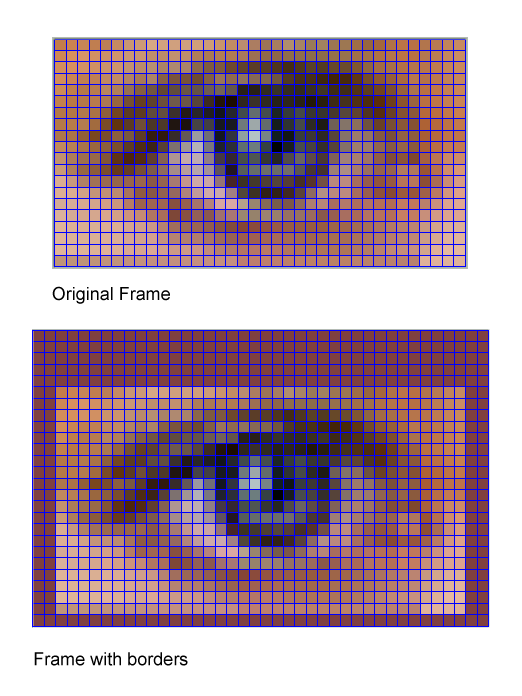 This class will enable you to add borders around the frames.
This class will enable you to add borders around the frames.
This can be useful if part of the information you want to segment is outside the field of view. Adding a border enable you to extrapolate outside the frame.
For more information, please consult the sliceOmatic 5.0 Online User's Manual
Class: Clone
 This module was developed by TomoVision for Yajima Nobuhiko of the Image Labo in Japan.
This module was developed by TomoVision for Yajima Nobuhiko of the Image Labo in Japan.
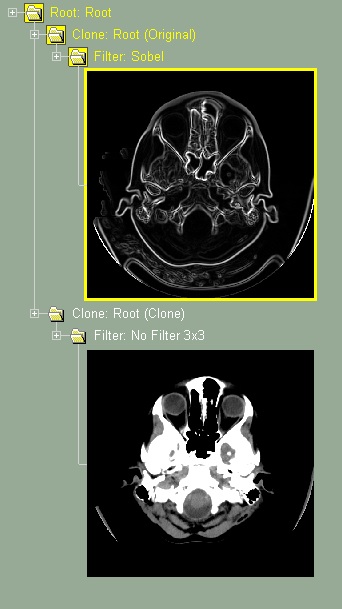
This class will create 2 branches in the database tree. One branch contain the original images, the other an exact clone of these images. Different treatment can be applied on the GLI images of each branch, but the segmentation TAG images will be identical in both the original and the cloned branches.
This class can be useful to compare the results of other classes that modify the GLI images (such as "Filter" or "Gradient"). For example, if you create a tree with the classes: Root / Clone / Filter / Frame, you can then apply different filters to the original and clone branches and compare the results. With that tree, the shortcut key "Insert" / "Delete" will toggle between the original and cloned images.
For more information, please consult the sliceOmatic 5.0 Online User's Manual
Class: Gradient Amplification
 This module was developed by TomoVision for the LIO group of the ETS in Montreal, Canada.
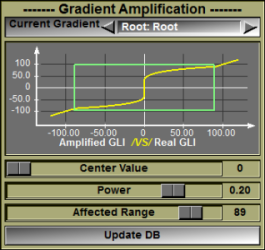
This module was developed by TomoVision for the LIO group of the ETS in Montreal, Canada.
This class enable you to artificially increase the gradient around a specific target GLI value. This will help segmentation techniques that are gradient based (such as "Morpho" and "Snake"). It can be used for example in segmenting CT images where you know you want to segment at specific Hounsfield values but do not want to use threshold based techniques. By increasing the gradient at the desired Hounsfield value, you insure that the gradient based technique will find the correct boundary.
For more information, please consult the sliceOmatic 5.0 Online User's Manual
Class: RR2D (Rigid Registration 2D)
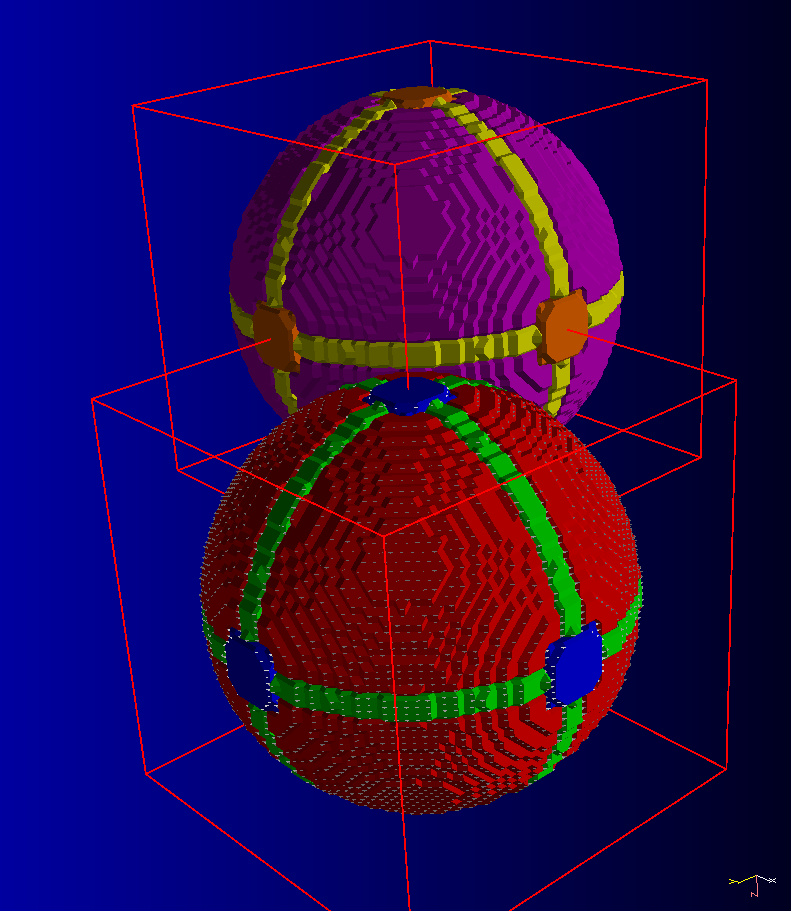
 This class will enable you to register slices in 2D (in the plane of the slices).
It can be used, for example, to re-align series if you need to move a patient between acquisitions.
This is a landmark based registration technique.
This class will enable you to register slices in 2D (in the plane of the slices).
It can be used, for example, to re-align series if you need to move a patient between acquisitions.
This is a landmark based registration technique.
For more information, please consult the sliceOmatic 5.0 Online User's Manual
Class: RR3D (Rigid Registration 3D)

 This class will enable you to register series in 3D using rigid transformations (translation, rotation and scaling).
This is a landmark based registration technique.
This class will enable you to register series in 3D using rigid transformations (translation, rotation and scaling).
This is a landmark based registration technique.
For more information, please consult the sliceOmatic 5.0 Online User's Manual
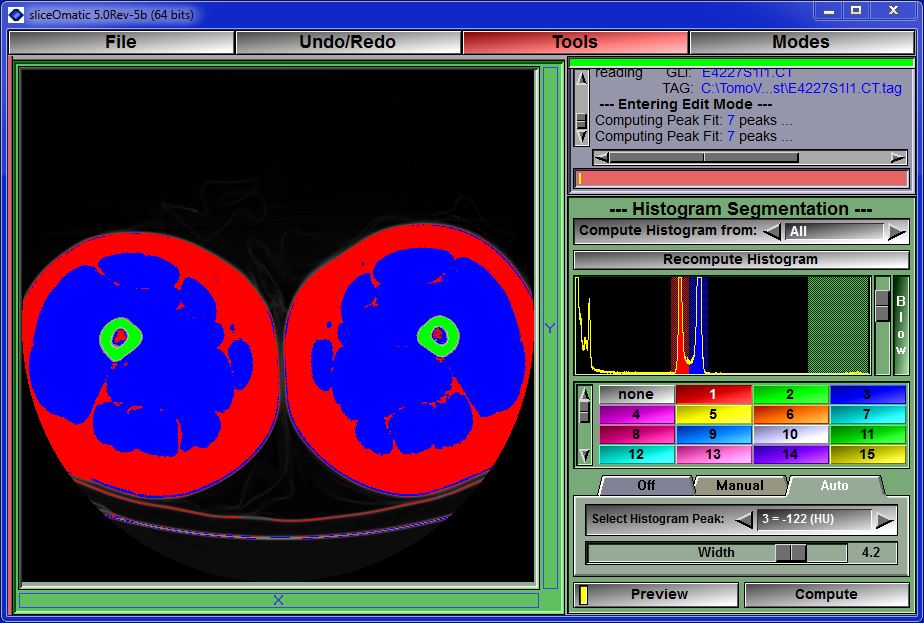
Mode Histogram Segmentation
 This module was developed by TomoVision for Jeffrey Cooley of McMurdoch University in Australia.
This module was developed by TomoVision for Jeffrey Cooley of McMurdoch University in Australia.
This mode is used to segment the images with the help of the histogram.
For more information, please consult the sliceOmatic 5.0 Online User's Manual
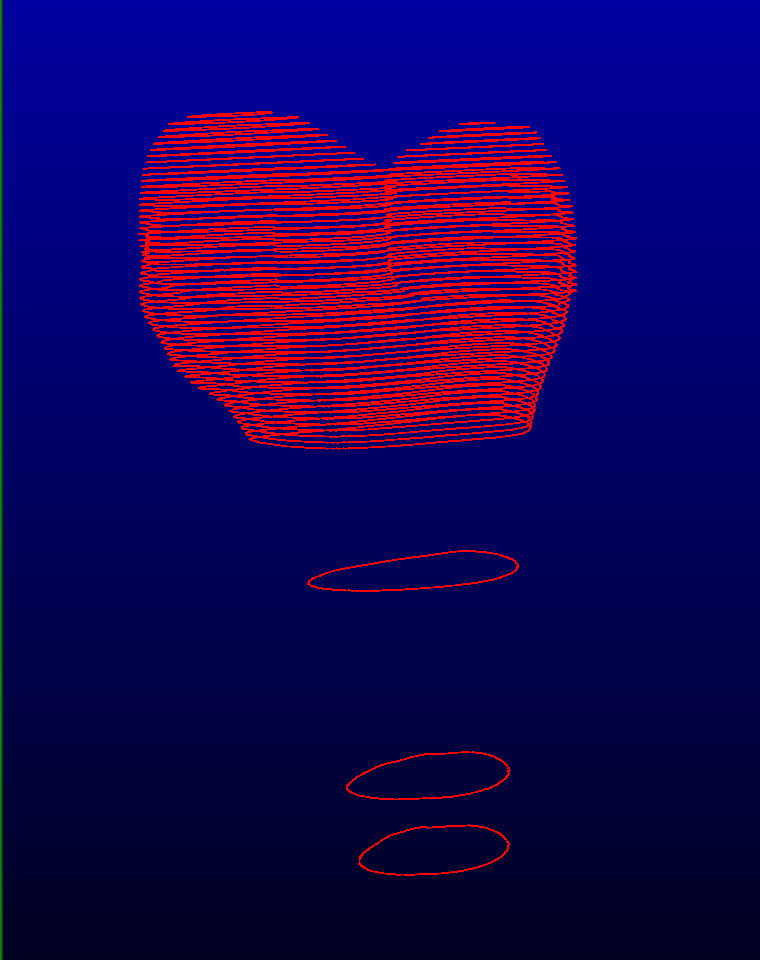
Mode TAG Interpolation
This mode is useful for 3D structures that do not change much between slices (ex: the long bones of the leg). You only need to segment a few slices and then let this mode interpolate the missing slices from the data on the existing slices.
For more information, please consult the sliceOmatic 5.0 Online User's Manual

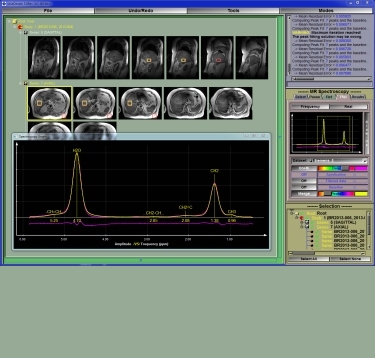
Mode MR Spectroscopy
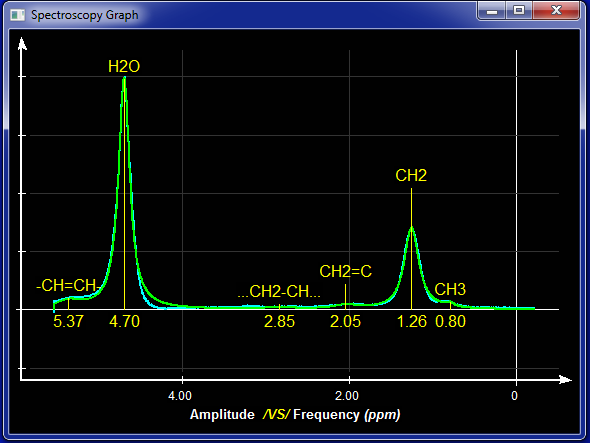 This module was developed by TomoVision for Robert Ross of Queens University in Kingston, Canada.
This module was developed by TomoVision for Robert Ross of Queens University in Kingston, Canada.
This mode enable you to analyse MR spectroscopy data.
Note:
At this time sliceO only recognise Siemens datasets. If you have any other kind of datasets, please contact us! We need sample data...
For more information, please consult the sliceOmatic 5.0 Online User's Manual
Required Hardware
SliceOmatic require a 64 bit Windows (either 7, 8, 10 or 11).
If you plan on analysing multiple slices or working with 3D volumes, we suggest having a lot of RAM memory (32Gb to 64Gb).
Starting with sliceO-6 revision 12 you can run sliceOmatic on the ARM version of Windows (the Surface Pro) and on the Parallels Windows emulator on a MAC system.
AI:
The AI modules need you to install Python on your system. Python is free and can be installed following these steps:
Python install
The AI modules are greatly accelerated (up to 10 times faster) if you have a CUDA compatible GPU on your system. Preferably a card with a lot of memory (3080 or better).
Ultrasound:
The ultrasound modules are written using the CUDA language, enabling them to use the power of the massively parallel Nvidia GPU.
But this mean that they can only work with graphic cards that have the Nvidia chipset. And even then, you need a
graphic card that support CUDA 8.0 or higher. This limit you to cards having the Ampere or newer micro-architecture.
To know if your graphic card is compatible you can check Nvidia's web site at:
https://developer.nvidia.com/cuda-gpus
VR:
The "VR Edit" and "VR Volume" ultrasound modes can only be used if you have Oculus gogles and touch devices installed on your system.
Security
I do understand that many hospitals have worries before installing a software that can access patient data on their system.
So, here's a few information on sliceOmatic that, I hope, will answer the questions you may have.
Data sharing: SliceOmatic does not communicate with any other devices, database, the cloud or TomoVision. It does not store any information outside itself. It does not even talk to the PACS! (All the images it analyze must be first copied from the PACS to the local PC).
The only patient information it access is the one present in the original DICOM files, and these are not copied or kept in any database. When you close the application, no data is retain anywhere.
Exported files: The program create 3 types of files derived from the original images:
- Segmentation file (.tag) that will have a map of the segmentation. These files only contain pixel information and no patient data. By default, these files are saved in the same location as the original DICOM files. So, either locally on your PC or on a shared drive accessible by Windows.
- Result file (.csv) that contain the surfaces/volumes computed from the original files. These file can contain patient information if these info have been selected for saving. The content and location of these results files are under the user's control.
- AI "Weight" files. If, to automate part of the segmentation process, the user train an AI on some images the program can save the computed AI weights. These files do not contain any patient information. They are saved locally and are not shared with TomoVision.
Anonymous mode: If you are doing a "blind" study, sliceOmatic has an "Anonymous" mode that can be enabled to prevent the user from seeing any patient information or saving it to the results files.