This group will have the buttons to access the interface of the 2D classes that do have an interface. By default, none of the active clases have an interface and this menu group is not available.
Activating 2D classes is done through the "Class Management (2D)" mode. Please note that only some of the classes have an interface (such as MPR, Mixer or Filters). The default classes (Study and Series) do not have an interface. Only classes with an interface will have a button in this menu group when they are activated.
 The some class can be activate more than once (for example, we can replace the Study and Series classes buy 2 instances of the DICOM tree class)
The some class can be activate more than once (for example, we can replace the Study and Series classes buy 2 instances of the DICOM tree class)
2D slices are sorted in a "class" tree. The different level in that tree are used to sort or modify the 2D frames.
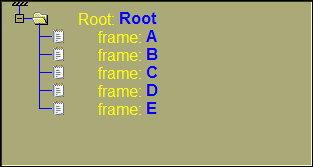
The 2D tree starts at the "Root" node, and finishes with "Frames" nodes. Each file that you read in sliceOmatic contains one or, sometimes, multiple images. Each images is a "frame". The root and the frames are both a "class" of nodes.
The root and the frames are always present in the database tree. But in between, you can add other classes.
Some of the classes, such as "Study", "Series" and "DICOM Tree" are used to sort the tree in patient/study/series/image hierarchy.
For example if we load 5 files in sliceOmatic, each having 1 image and with the following parameters:
|
|
Image files |
Study ID |
Series number |
|
|
A |
0001 |
1 |
|
|
B |
0001 |
2 |
|
|
C |
0002 |
1 |
|
|
D |
0002 |
1 |
|
|
E |
0002 |
2 |
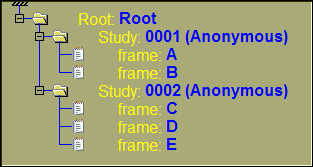
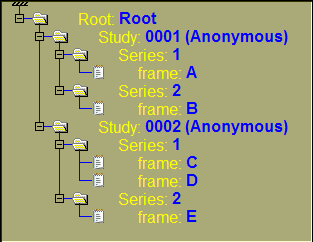
If we create different combinations of trees using the Study and Series classes on top of the default Root and Frames, we will have the following trees:
|
|
|
|
|
|
|
|
Root & Frame |
+ Study |
+ Series |
+ Study & Series |
By default, the tree contains the root, study, series and the frame classes. This is to create a tree that is similar to the one present in sliceOmatic 4.3.
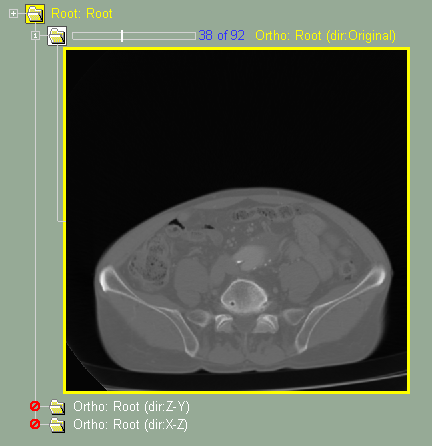
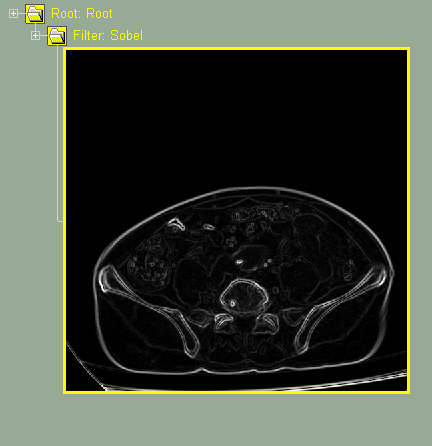
Other classes, such as "Filter Conv", "MPR Ortho" and "ROI", are used to modify the GLI images.
|
|
|
|
|
|
|
|
Original Frame |
Filter |
MPR |
ROI |
The classes that can have an interface buttons are:
Add a level of the class tree used to display the images in sliceO. You can select the DICOM tag used to discriminate the images in that tree level. For example, if you use the DICOM tag (0020,0010) = Study ID, then this class is the equivalent to the "Study" class.
This class enable you to change the sort order of the images.
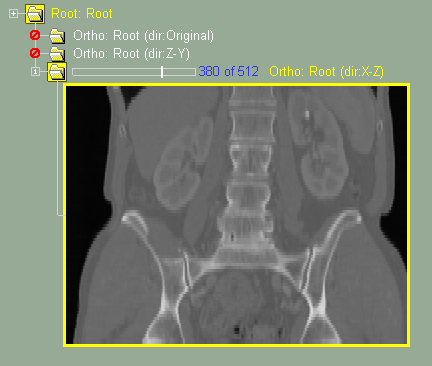
This class enable you to create new images from an original 3D dataset.
This class enable you to create new images from an original 3D dataset.
Note:
|
|
|
This class enable you to create a scout images from an original 3D dataset.
Note:
•ROI
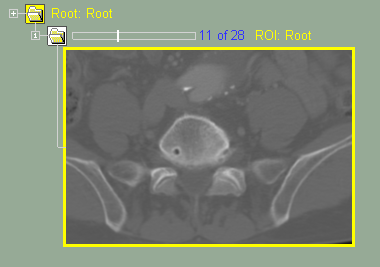
This class enable you to select a portion of the original images to work with.
This class enable you to mix images together. It can be used with DIXON or IDEAL MR images to take advantage of all the images of the same slices.
This class enable you to work with multiple images of the same slice.
This class enable you to filter the images using convolution filters.
This class enable you to filter the images using FFT filters.
The classes that do not have an interface buttons are:
If this class is placed in the DB tree, there will be one instance of this class per study.
If this class is placed in the DB tree, there will be one instance of this class per series.
If this class is placed in the DB tree, there will be one instance of this class per acquisition.
•Path
If this class is placed in the DB tree, there will be one instance of this class per different path of the TAG files.
If this class is placed in the DB tree, there will be one instance of this class per different suffix used in the TAG files.
The optional classes for this group are:
This class enable you to add borders to the images.
This class create a second copy of the images.
This class change the GLI values of the images according to their histogram.
This class enable you to create new images of a 3D group.
•RR2D
This class enable you to align 2D images.
•RR3D
This class enable you to align 3D groups of images.
Note:
|
|
|
From the Display Area
The mouse controls associated with this classes are:
|
|
|
|
|
|
Mouse button |
Function |
|
|
|
|
|
|
Scroll Wheel |
Increase / decrease the magnification of the selected image(s). |