In this step, you will read the files needed to select and find the target slices of the thigh into the sliceOmatic program.
The Interface:
What the script did for you:
The "DB File Management (2D)" mode interface is open.
Only window 1 is visible and it is in "Mode ALL".
The "Color Scheme" tool is open.
The color scheme is set to "Mix".
The "DICOM Browser" is open.
What to do:
|
|
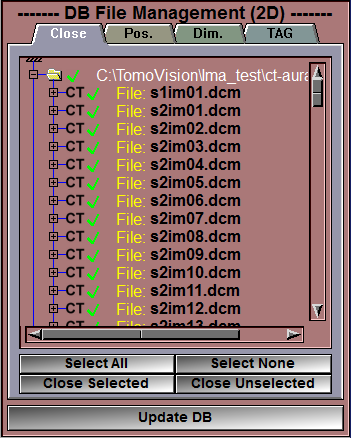
First, if you have any previously loaded files, you can close them using the "DB File Management" interface.
•First, click on the "Select All" button
•Then, click on the "Close Selected" button
•Finally, click on the "Update DB" button |
|
|
|
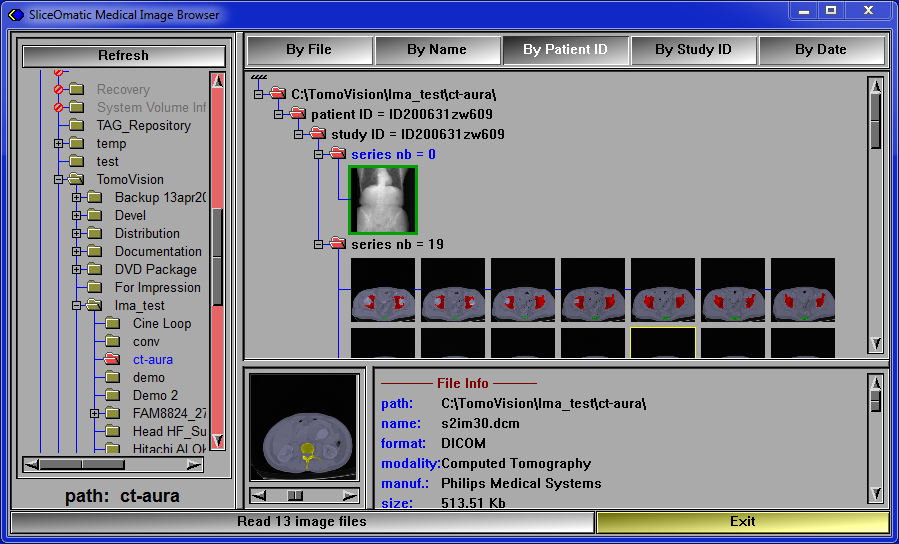
You can use the DICOM Browser to select the study you will analyze. |
|
|
|
You can find step by step instructions by following this link.
|
|
Or, you can simply drag & drop the desired files directly on the sliceOmatic's window.
Note:
|
|
The Baylor Thigh Protocol use IDEAL or DIXON files, this mean that you need both the Water and Fat (or Water Suppressed) slices of the thigh. |
|
Note:
|
|
To be able to select the desired axial slice from an image in a different view (scout, coronal or sagittal) in the next step, all the images must be in the same frame of reference (DICOM tag (0020,0052)). This is usually the case for all the series inside the same study but not always.
Using the sorting "By Study ID" will sort the images by study ID, but also by frame of reference. You can use this sorting mode to insure that all the loaded files are within the same frame of reference. |
All the read files will be visible in the sliceOmatic's window.
|
|
You can decrease/increase the size of the displayed images with the "+" and "-" keys on the keyboard.
|
|