In this mode, you can use an AI to segment your 3D volumes, or if you want, you can train an AI on your previously segmented volumes.
This module work with 3D stack of images to automatically compute their associated TAG values.
The program will reslice the volumes along the "z" axis and work with the reslice data. So, your input volumes do not need to be in any specific orientation, you can use axial, coronal or saggital slices. You can have a gantry tilt. You can even work with volumes expressed in a spherical or cylindrical system.
The volumes will be resliced using the pixel size you specified in the Config/PreProc page. So, your input volumes can have any pixel dimensions you want. The re-slice volume will have isotropic voxels (same dimensions in X, Y and Z).
For the 3D module, we introduce the concept of "Patches". 3D volumes are to big to fit in memory, so we will work on a subset of the complete volume: the patches. The volume will be subdivided in multiple cubic patches and the AI will be trained on these patches instead of directly on the volumes. You will have control over the patches in the "Preproc" tab of the configuration pages.
Subdividing the volume in patches may introduce artifacts at the border between the patches. To prevent this, you can specify an "overlap" between the patches. When predicting TAG values we will discard the voxels on the perimeter of each patch.
Note:
|
|
|
From the Graphic Interface
|
|
|
|
|
||
|
|
The main "AI with Python" interface has 3 tabs: Train, Test and Predict. Each of theses has a "Config" button that will open the configuration menu, and one (or more) "Compute" button that will open the appropriate page. |
||||
The Configuration Menu
Each of the 3 main pages of the "AI Python" interface has a "Config" button. This will open the configuration menu. Depending on the main pages you are in, the configuration menu will have between 3 and 8 sub-pages. For training, you need access to all the pages, for "Predict", you only need 3.
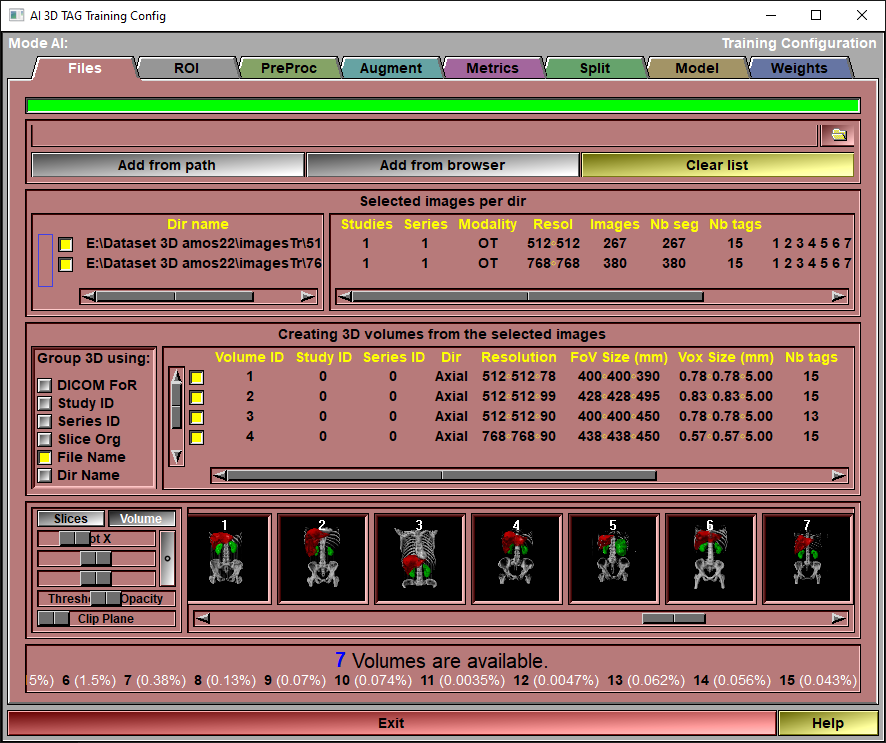
The config "File" page
This page is used to specify the images that will be used either for training or testing your AI.
|
|
|
||||||||||
Progress bar |
Show the progress of the program when parsing directories
|
||||||||||
Add slices |
Files can be added to this list in 3 different ways:
Simply drag&drop a directory on the interface. The program will recursively parse this directory and all its sub-directories. Each directories that contain segmented images will be add as a separate line in the interface or use the "From Path" or "From Browser" options.
|
||||||||||
From Path |
You can achieve the same results by specifying the directory in the input line and clicking the "Add from path" button.
|
||||||||||
From Browser |
You can add files with the "Medi Browser" interface.
|
||||||||||
Clear List |
Clear the list.
|
||||||||||
Dir List |
For each directory specified that contain segmented files, you will see a line of information specifying the number of studies in that directory, the number of series, the modality of the images in the directory, the resolution of these images, the total number of images and the total number of segmented images, the number of TAGs present in these segmented images and a list of these TAGs.
You can also select and de-select any of the directories by clicking on the associated button in front of each line.
Please note that only the segmented images (those that have an associated ".tag" file) are used. If a directory does not contain any segmented images, it will not have a line in the interface.
A double click on a line in this list will open the associated files in sliceOmatic.
|
||||||||||
Group 3D Using |
Normally, using the DICOM Frame of Reference (FoR) UID is enough to differentiate volumes. But if that info is not available (for example with anonymized data), you can use one of the other parameters to differentiate the volumes: Study ID, Series ID, or the x/y slice origin (usually all the slices in a 3D stack have the same x and y origins), file names or files directory.
|
||||||||||
Volume List |
A list of the different identified volumes. Some info is provide about each volumes. If one of the parameters is a red "Mix" value, this mean that the volumes have not been differentiated properly.
The volumes can also be disabled from this using the check-mark on the left hand side of the list.
|
||||||||||
Volume preview
|
A preview of all the selected volumes is available. Volumes can be seen as either a series of 2D slices (in any of the 3 main directions) or as ray-traced volumes. When viewing volumes, you can adjust the threshold slider and place a clip plane. The threshold slider will have different functions depending on the current display modes (display modes can be changed with the "Color Scheme" tool or with the F1 to F4 keys.
|
||||||||||
Files status |
Show the number of volumes found in the different selected directories. Also, the different TAGs found in these volumes (along with their prevalence) will be displayed.
|
||||||||||
Exit |
Exit the configuration interface.
|
||||||||||
Help |
Display this web page. |
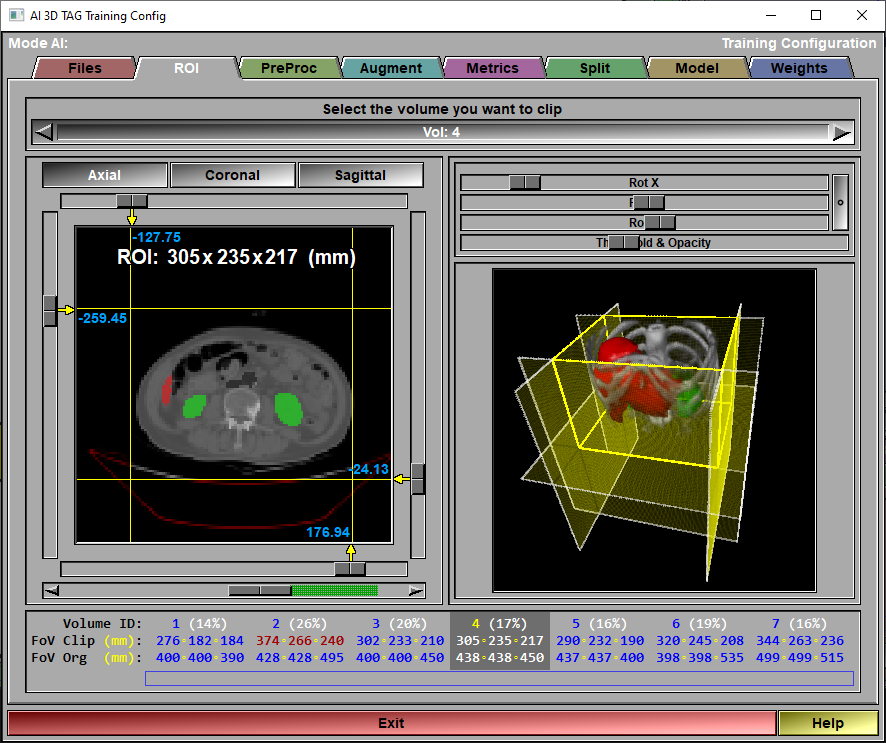
The config "ROI" page
This page is used to limit the size of the volumes. You can use clip planes to define a Region of Interest (ROI), removing parts of the volumes that do not contribute to the AI training.
These "clipping" values will automatically be saved in a file in the same location as the original slices. This clipping information will have the same name as the first slice in the volume (if the volume is composed of individual slices) or of the volume (if the volume is from a 3D file) with the "3D_clip" extension. The file is a simple ASCII file that can be looked at with notepad. It contain 3 lines for the 3 x,y,z axis. Each line give the positions (in mm) of the 2 clip planes on this axis.
|
|
|
Note:
|
|
|
|
|
|
Volume selection
|
Use this tool to select the volume you want to clip
|
ROI window
|
the clip planes can be adjusted with the left/right/top and bottom sliders. Change the view from Axial to Coronal or Sagittal to access the clip planes in the other axis. The slider at the bottom is used to scroll through the slices of the volume.
|
Volume view |
A 3D ray-traced view of the volume is also available.
|
Volume status |
Information on each of the loaded volumes will be displayed here.
The information is presented on 3 lines:- The Volume ID (defined by the order the volumes where loaded in the AI module) along with the percentage of the original volume present in the ROI.- The original FoV of the volume- The clipped ROI dimensions. If no clip planes are defined, the ROI is the same as the original FoV.
The background of the current volume is presented in a darker grey than the others.You can select the current volume by clicking on it's values in this windowThe color of the numerical values have a meaning: Red is used for the maximim value in any of the 3 directions.
|
Exit |
Exit the configuration interface.
|
Help |
Display this web page. |
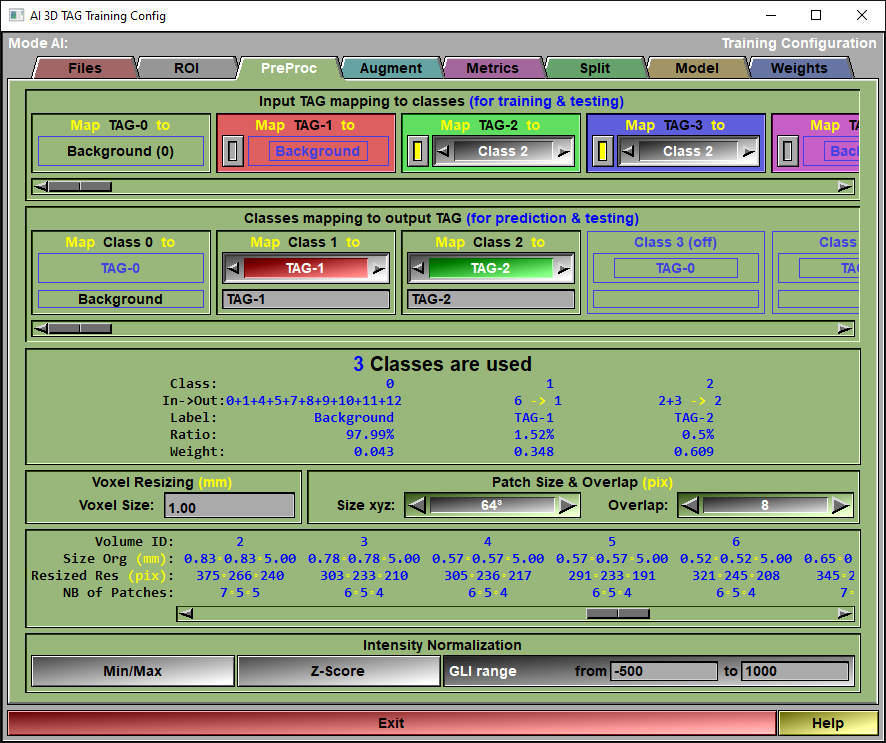
The config "Preproc" page
This page is used to control any pre-processing that need to be applied to the volumes. You also use it to select the classes used in your AI.
Note:
|
|
|
|
|
|
Input TAG mapping
|
In this section, there is one box for each TAG present in the selected images. The title of the box (ex: "Map TAG-1 to") let you know what TAG is being mapped. Each of these can be mapped to a "Class" for the AI.
At first, there is as many classes as there are TAGs in the selected images. But since multiple TAGs can be mapped to the save class, some of the classes can be disabled if no TAG mapped to them. So, if there is a TAG in the images you do not want to use in the AI, just map it to the background.
This mapping is only used for training and testing. When predicting, only the grey values of the image is used.
|
Class mapping
|
Each classes used in the AI can be mapped to a TAG value. So, for example, if you map "Class 1" to the "TAG-1", when predicting the segmentation of an image, the tissue associated to Class 1 will be tagged in sliceO using TAG-1. You can also assign a label to that TAG. That label will be used in the AI reports and, if you use the AI to segment images, assigned to the corresponding TAG in the sliceO's interface.
This mapping is only used for predicting and testing.
|
Mapping status |
An overview of the current classes and how they are mapped, along with their label and the ratio of pixels (with the associated weights) is presented here. The "Weight" values are used in the different "Weighted" metrics.
|
Voxel Resizing |
The 3D AI need the voxels of all the volumes to be isotropic and of the same size. You specify the desired voxel size here and sliceO will take care of re-sizing all the volumes.
|
Patch Size & Overlap |
Chances are your computer does not have enough memory to train an AI using the complete volumes. So, we will split the volumes in smaller "patches". You must define the size of these patches. Patches are cubic with x,y and z having the same dimentions. You can also specify an overlap for the patches.
|
Patch status |
This window present you with the new resized resolution and number of patches for each volumes.
|
Intensity Normalization |
The AI expect floating point pixels values between 0 and 1. While medical images use integer values. So, we need to "normalize" the pixel values.
You have 3 choices: "Min/Max", "Z-Score" or "GLI range".
The "Min/Max" normalization will normalize the GLI values so that the minimum value found in an image will be 0 and the maximum will be 1.
The "Z-Score" normalization will normalize the GLI values so that the mean value of an image will be at 0.5 and and the standard deviation is 0.5. The idea is that all the values within 1 standard deviation of the mean will be between 0 and 1.
The "GLI range" is mainly used for CT images. All HU values under the minimum will be assigned a value of 0, and all HU greater than the maximum will be assigned a value of +1 in the normalized images. All HU values in-between will be linearly mapped between 0 and 1.
|
Exit |
Exit the configuration interface.
|
Help |
Display this web page. |
Note:
|
|
|
Note:
|
|
|
Note:
|
|
|
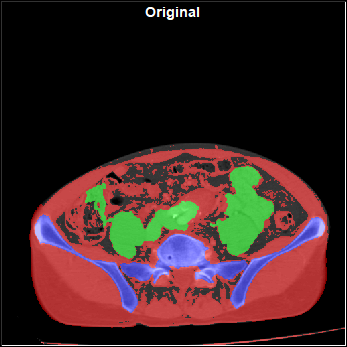
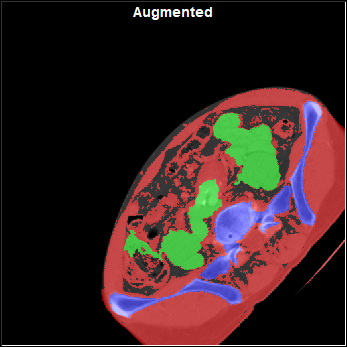
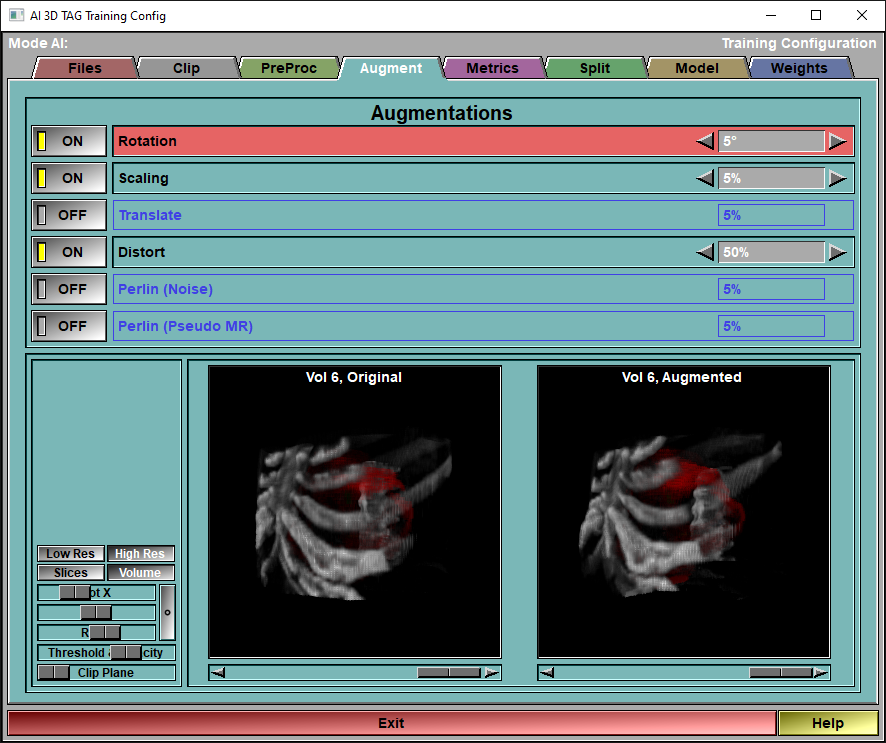
The config "Augment" page
This page enable you to select and control the different augmentations that are applied to the images before they are used for training. The augmentations will be applied to all the "training" images before each epoch. Only the "Training" images are augmented, the images reserved for validation and testing are never augmented.
|
|
|
|
|
|
|
|
Original Image |
Rotated |
Scaled |
Translated |
|
|
|
|
|
|
|
Distorted |
With Noise |
With "Pseudo MR" |
|
|
|
||||||||||||
Augmentations |
You can enable/disable the different augmentations, and specify the intensity of each of these from this interface.You have access to 9 augmentations. Each of these is controlled by a selected value "val":
|
||||||||||||
Demo Original |
The slider under the image is used to change the demo frame. All the volumes used for training are available.
|
||||||||||||
Demo Augmented |
The slider under the image is used to show the range of augmentation.
In normal training images, each time an image is augmented, a random value between -1 to +1 is assigned to each augmentation of each image.
In this demo however, we use the slider to show the range of possible augmentation. The slider simulate the random variable, with a range of -1 (complete left) to +1 (complete right).
|
||||||||||||
Exit |
Exit the configuration interface.
|
||||||||||||
Help |
Display this web page. |
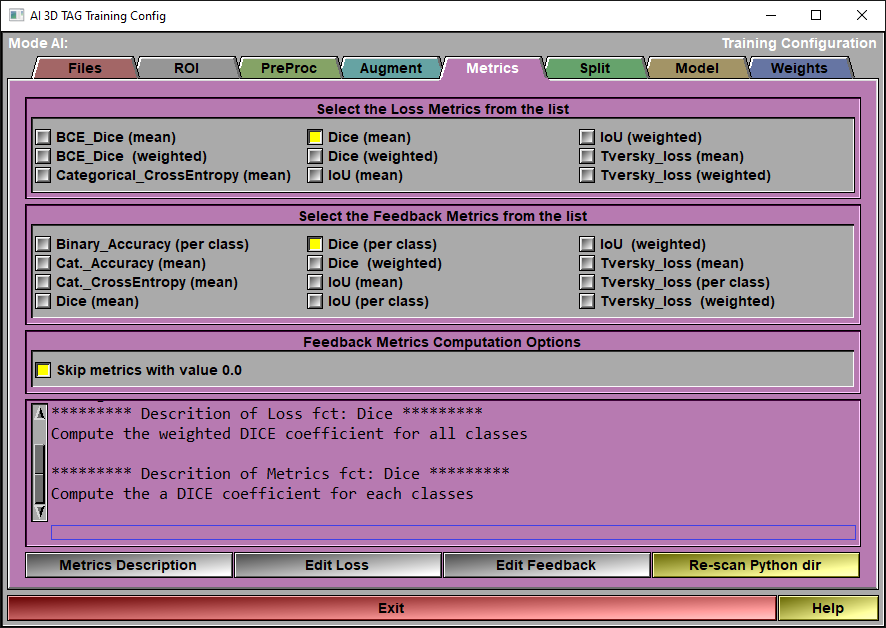
The config "Metrics" page
When you train the AI, 2 sets of "metrics" are used: The Loss Metrics is the one used to compute the AI weights. The other group of metrics are just computed to give you an idea of what is going on. They are only there to provide you some feedback on the accuracy of the AI, they are optional and can be all off if you want.
At this time, you can only have 1 "Loss" metric when training your AI. You can however have as many "Feedback" metrics as you want.
These metrics will be reported back to you when you train your AI. They can also be used when you will select the weights you want to save for your AI. During training you will be presented with 2 graphs: One for the training and validation loss, and one for the training and validation values of the feedback metrics.
|
|
|
Loss Metrics |
Select one of the metrics as a "Loss" function to train the AI.
|
Feedback Metrics |
Select any metrics you want from this list to get additional information on the AI training progress.
|
Computation options |
A lot of the patches do not contain any voxels for at least one of the classes use in training. If the predicted value for the same patch contain one or more voxels of that class, some of the metrics (such as Dice or Jaccard) will have a resulting value of "0". All these "0" values cause by single voxels skew the mean value of the metrics. You can select to disregard these "0" values in the mean computation. Be advised that although this look like a good deal, the absence of TAG data is an information that the training can use. If you remove all empty patches, the prediction may not know what to do in these regions later on.
|
Text Window |
This window reports on the metrics selections and the description of each metrics.
|
Description |
This will cause the description associated with each of the selected metrics to be displayed in the text window.
|
Edit Loss |
The different metrics are all Python files that you can edit if you want. You can also add your own metrics. Just follow the prescribed syntax and add the python file of your new metric in the Python directory. Then click the "re-scan dir" to cause sliceO to re-analyze the directory and add your metrics. The syntax will be described later at the end of this section. |
Edit Feedback |
|
Re-scan dir
|
|
Exit |
Exit the configuration interface.
|
Help |
Display this web page. |
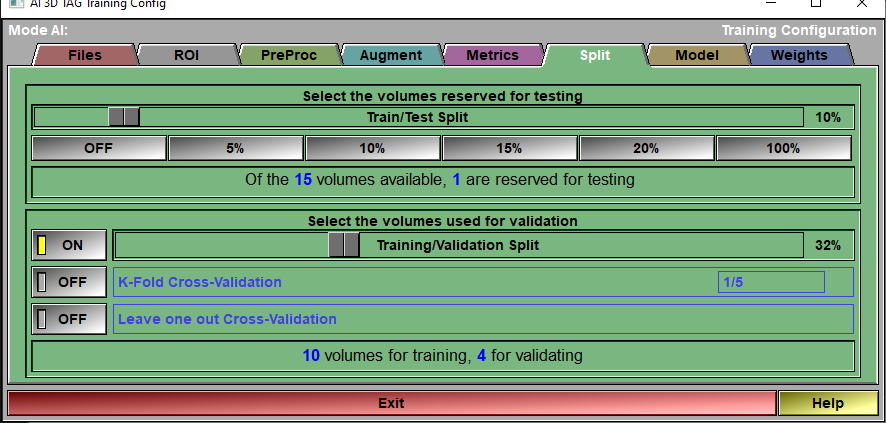
The config "Split" page
|
|
|
Testing |
A portion of the volumes you selected in the config "Preproc" page can be reserved for testing. If you do not want to do any testing with the loaded volumes, you can set this slider to 0% with the OFF button. If on the other hand you loaded volumes for testing only, set the slider to 100%.
|
Testing Status |
Report on the number of available volumes
|
Validation |
You have access to 3 validation methods:•Training/Validation Split. Probably the simplest, a part of the available volumes (those left after the volumes reserved for testing are removed) is reserved for validating the AI at each epoch. The same volumes are used for validation at each epoch. •K-Fold Cross-Validation. The available volumes are split in "n" groups using the ratio defined in the K-Fold interface. For example if the K-Fold is 1/5, then n=5. We do a complete training of all the epochs reserving the volumes of one of these groups validation. We then redo the complete training using another group for validation. We do this n times. After that, we assume that we have a good idea of the validation since by now all the volumes have been used for that, we do a last training without any validation. In the "Train Save" page, the graphs will show the mean value of the n+1 pass for training metrics and the n pass with validation for validation metrics. •Leave one out Cross-Validation. This is the extreme case of K-Fold where n is equal to the number of volumes.
|
Validation Status |
Report the number of volumes that will be used for training and for validation.
|
Exit |
Exit the configuration interface.
|
Help |
Display this web page. |
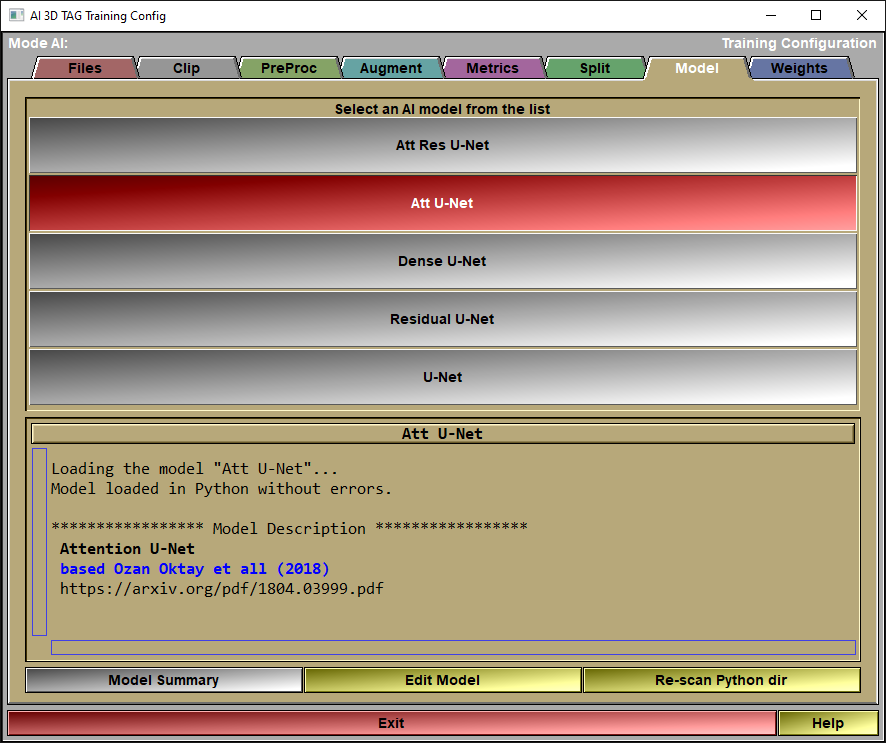
The config "Model" page
In this page, you will select the model you want to use for the AI.
|
|
|
Model List |
Select a model from the list.
|
Model Feedback |
Information on the selected model will be displayed here.
|
Model Summary |
Call the Keras function "Summary" for the selected model.
|
Edit Model |
The different models are all Python files that you can edit if you want. You can also add your own models. Just follow the prescribed syntax and add the python file of your new model in the Python directory. Then click the "re-scan dir" to cause sliceO to re-analyze the directory and add your model. The syntax will be described later at the end of this section.
|
Re-scan Python dir
|
|
Exit |
Exit the configuration interface.
|
Help |
Display this web page. |
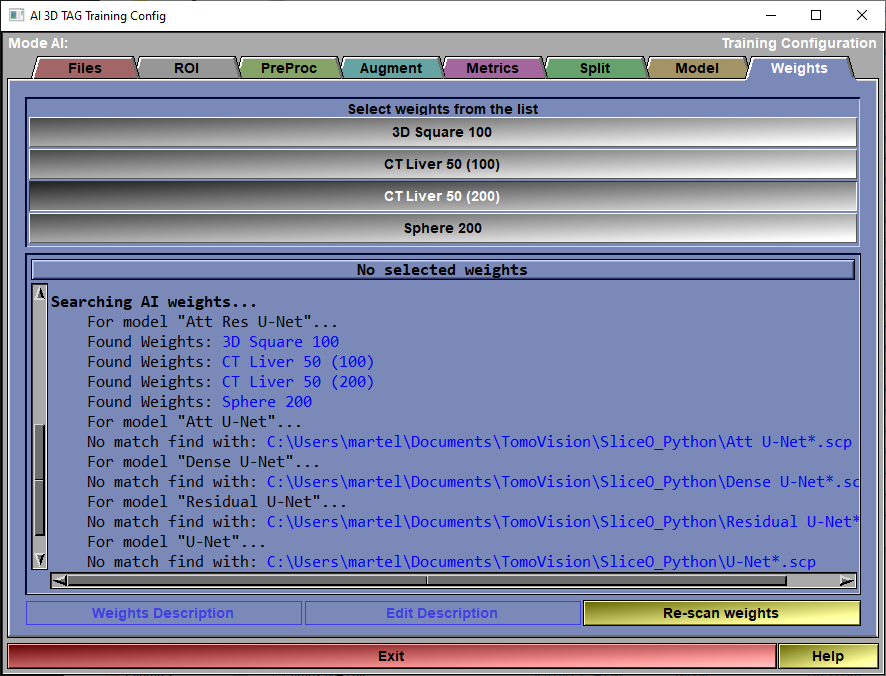
The config "Weights" page
|
|
|
Weight List |
This list will display all the available weights for the selected model. Select one of these from the list.
|
Weight Feedback |
Information on the selected weights will be displayed here.
|
Weight Description |
Cause the description of the weights to be displayed in the text window.
|
Edit Description |
The weight description is a text file associated with the weights. It has the same name as the weights but with the ".txt" extension. It can be edited with a simple text editor. If you click "Edit", this file will be opened in Notepad.
|
Re-Scan weights
|
Re-scan the directory if you have saved some new weights for this model. |
Exit |
Exit the configuration interface.
|
Help |
Display this web page. |
|
|
|
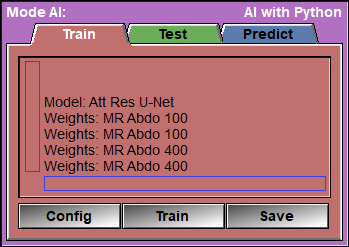
Text Feedback |
|
Config |
Open the Configuration window
|
Train
|
Open the "Train" Window. |
Save |
Open the "Save" Window.
|
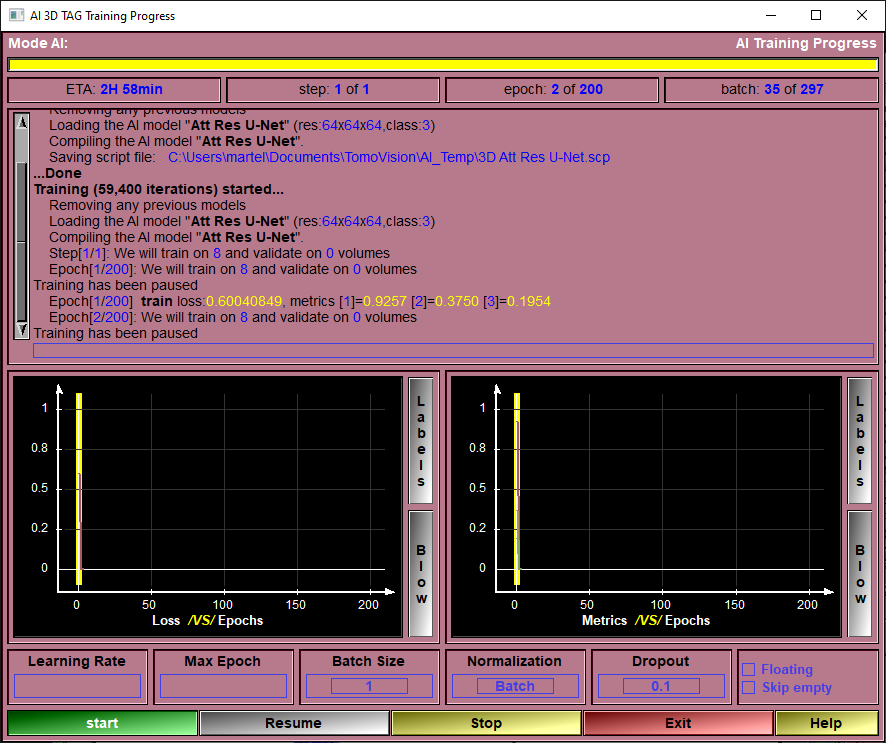
Once you used the config pages to select the files you want to train on, and you selected the preprocessing steps, the augmentations, the validation split and the model you want to use, you are ready to start training your model to obtain the AI weights.
This is the page that will enable you to train the AI.
The only things that remain for you to select are the learning rate, the number of epochs and the batch size. After that, you click on "Train" and wait (and wait... and wait...) for the results!
Once started, the training will for each step:
•Reset or pre-load the weights (if you selected to re-train existing weights).
Then, for each epoch it will:
•Make a scrambled list of the volumes used for training.
Then for each of the training volumes in that list:
•Split the volume in patches.
•Load, normalize and augment the next "batch" of patches.
•Call the Keras function "train_on_batch" for this batch of patches.
Then for each validation volumes:
•Split the volume in patches.
•Load, normalize the next "batch" of patches.
•Call the Keras function "test_on_batch" for the batches of patches of the validation volumes.
•Report the results of this epoch and possibly save the current weights, the current metrics and a script associated with these to the "AI Temp" folder. We only save a total of 100 weights, so if you have more than 100 eopchs, we will only save weights each (nb_epoch/100) epochs.
The number of "steps" is dependant on the validation technique. A simple Train/Validation split us done in a single step. K-Fold and Leave-one-out cross validation require multiple steps.
Note:
|
|
|
Note:
|
|
|
Note:
|
|
|
|
|
|
Progress Bar |
Report on the training progression.
|
ETA |
Estimated time of completion (computed after each epoch).
|
Step Feedback |
Current step number.
|
Epoch Feedback |
Current Epoch number.
|
Batch Feedback |
Currently processed batch.
|
Text Feedback |
textual information about the training progression.
|
Loss Graphic |
A graphic showing the training and validation loss values as a function of the epochs.
|
Validation Graphic |
A graphic showing the training and validation values of all the selected feedback metrics
|
Learning Rate |
The learning rate used in training the AI. the default value is 0.001
|
Max Epoch |
The number of epoch used for the training.
|
Batch Size |
The number of images used in each training and testing batchs. If the AI failed becuase of a lack of GPU memory, you can decrease this value.
|
Normalization |
The AI need the values of the training dataset to be close to the range "-1" to "1". But a lot of the model's computations will cause the values to drift from this range. You can select to "Normalize" the data at each step of the model. You have the choice of 3 normalization modes: •Batch •Group •Instance
The "Normalization" value is placed in the system variable: "$AI_NORMALIZATION" and is accessible to the model. This is explained further down.
|
Dropout |
A technique to help the training to converge is to "drop" a number of the computed weights at each step of the computation. A "dropout" of 0.3 for example will reset 30% of the weights to zero. This help prevent the training from converging toward a false local minima.
The "Dropout" value is placed in the system variable: "$AI_DROPOUT" and is accessible to the model. This is explained further down.
|
Floating |
When a volume is "split" in patches, the patches cover a region that is always larger than the original volume. For example a 256x256x256 volume computed with a 48x48x48 patches will be subdivided using 6x6x6 patches. The patches cover 6X48 = 288 voxels. There will be a 16 voxel border around the original volume. If you enable the "floating" option, the origin of each patches will have a random offset of up to 16 voxels.
|
Skip empty |
Some patches do not contain any of the target classes (appart from class-0, the background). You may chose to disregard these patches when training.
|
Start |
Start the training.
|
Pause/Resume |
You can pause and resume the training.
|
Stop |
Stop the training.
|
Exit |
Exit the "Training" interface.
|
Help |
Display this web page. |
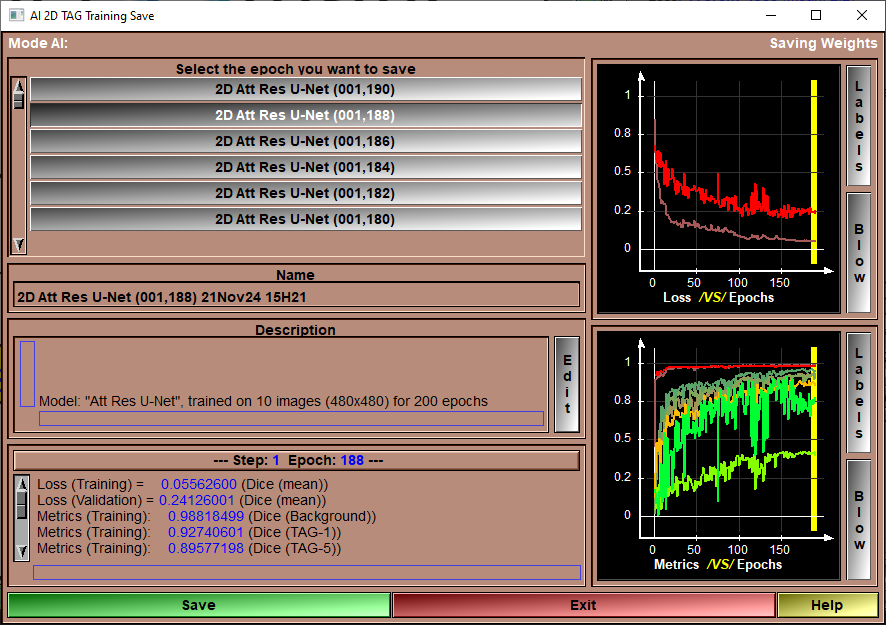
After having trained your AI, you want to save the weights you just computed.
This page will show you the weights of all the epochs of the training. You need to select one of these and save it.
When clicking "Save" 4 files will be transferd from the "AI Temp" folder to a sub-directory of the "Python Code" folder. This sub-folder will have the name of the model used for the training (ex: "U-Net", "Att Res U-Net"...). That sub-folder will contain 4 files, they will all have the name you selected but with different extensions:
•The Keras generated weights (with the extension ".data-00000-of-00001"). This file will be big (more than 100Mb).
•The Keras generated index (with the extension ".index"). This file has between 30Kb to 40Kb.
•A sliceOmatic script file containing the training parameters (with the extension ".scp").
•An ASCII "description" file (with the extension ".txt").
Note:
|
|
|
To help you select the optimal epoch for the weights, you have access to plots of the loss and feedback values along with their numerical values.
|
|
|
Epoch Selection |
Select the desired epoch from the list.
|
Name |
Name use for the saved weights. A default value is proposed, but you can change it as you which.
|
Description |
The description associated with the saved weights. Again, a default value is proposed, but you can edit this test to your liking.
|
Epoch Feedback |
Numerical values of the different metrics at the end of the selected epoch.
|
Loss Graphic |
Plot of the training and validation loss as a function of the epochs. The yellow vertical bar represent the selected epoch. You can also select the epoch to save by clicking at the desired location in the graphic window
|
Validation Graphic |
Plot of the training and validation feedback metrics as a function of the epochs. The yellow vertical bar represent the selected epoch. You can also select the epoch to save by clicking at the desired location in the graphic window
|
Save |
Save the selected weights to the model's sub-folder.
|
Exit |
Exit the "Save" interface.
|
Help |
Display this web page. |
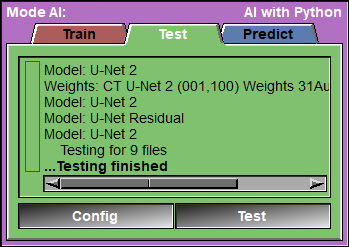
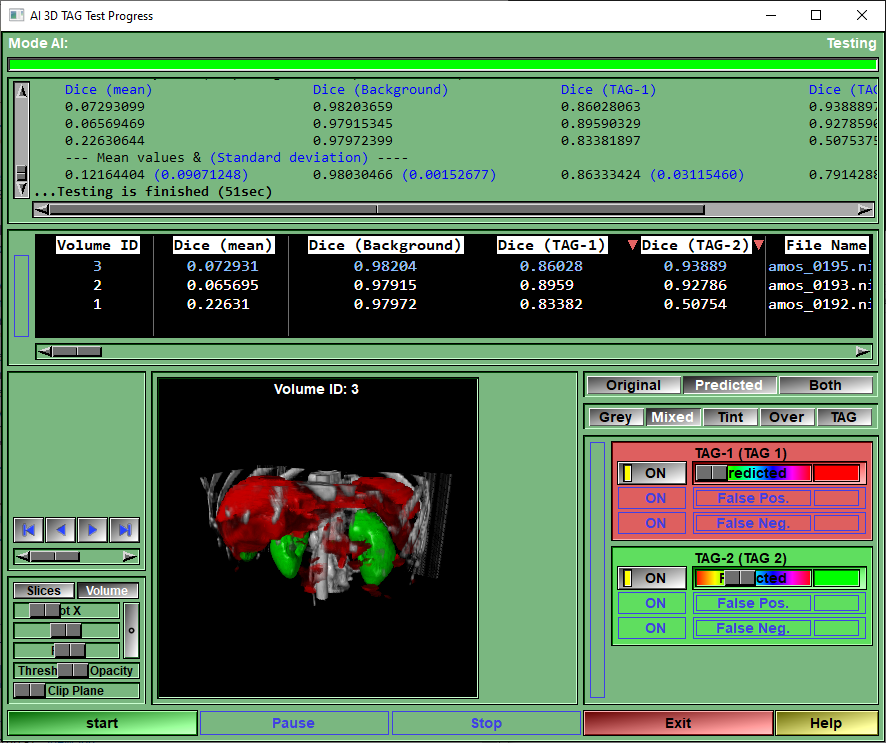
This page is used to test your AI against a reserved portion of the selected blocks.
The program will go through all the "test" blocks, and call the Keras "test_on_batch" function for each of them. It will report the different metrics for each blocks and once all the blocks are tested, compute the mean and standard deviation of each of the metrics.
These results will also be saved in a Excel compatible file in the AI_Temp directory.
|
|
|
Text Feedback |
|
Config |
Open the Configuartion window
|
Test |
Open the "Test Progress" Window.
|
The reconstruction is done "by patches", but some patches may have no TAG in the original patch and and only 1 in the predicted patch. This will give strange values for most metrics (Dice and Jaccard will both be 0) and skew the mean values of the metrics. This is why I do a first pass where I use the patches to compute the predicted TAG data (and display the results per patches), then a second pass where I reconstruct the complete volumes (both original and predicted) and recompute the metrics on theses. We get more significant values that way. The values presented in the "Result box" are the values for the complete volumes.
|
|
|
Progress Bar |
Show the progression of the testing
|
Text window |
Report on the metrics for each patches/volumes Also display the computed mean and variance values for these metrics.
In the first pass (computed with patches) the mean and variances will be for the patches of each volume. For the second pass (computed with volumes), the mean and variances are for all the volumes.
|
Results box
|
Once the testing is done, this box will display the numerical results of the different metrics computed from the complete volumes.
The results can be sorted according to any of the presented columns. Just click on the column title to change the sort order.
The line matching the test volume selected for display will be highlighted in blue. You can select any of the test volumes for display by clicking on its line.
|
Volume selection |
You can display any of the volumes used for testing. the arrow keys will advance to the first volume, the previous volume, the next volume or the last volume. You can also use the slider to select volumes. The sort order of the volumes is defined in the Results box.
|
Display controls |
The displayed volumes can be presented as a 3D volume or slice by slices.
When showing a volume, the sliders can be used to rotate it (it can also be rotate directly from the display window). The "o" button re-set the rotations. The "Threshold" slider will have different funstions depending on the "Display Mode". And the "Clip Plane" slider cna be used to control a clip plane parallel to the display window.
When showing the slices, the 3 sliders can be used to select the displayed slice in any of the 3 major axis.
|
Display window |
Display the test volumes.
|
Source selection |
You can display either the original segmentation, the predicted segmentation, or a mix of both. When using a mix of both, the false negative and false positive voxels will be brighter than the voxels that are a perfect match.
|
Display Mode |
This is the same controls as the "2D Color Scheme" tool in sliceOmatic. the "F1" to "F4" keys can also be used as shortcuts.
|
TAG Color |
For each TAG computed by the AI, you enable/disable and change the color of 3 group of pixels: •The pixels that have this TAG value in both the original and predicted images. •The pixels that have this TAG value in the predicted image only (False Positive) •The pixels that have this TAG value in the Original image only (False Negative)
|
Start |
Start the computation of the tests.
|
Pause/Resume |
Pause/resume the computation.
|
Stop |
Stop the computation.
|
Exit |
Exit this page.
|
Help |
Display this web page. |
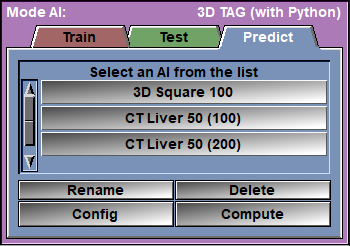
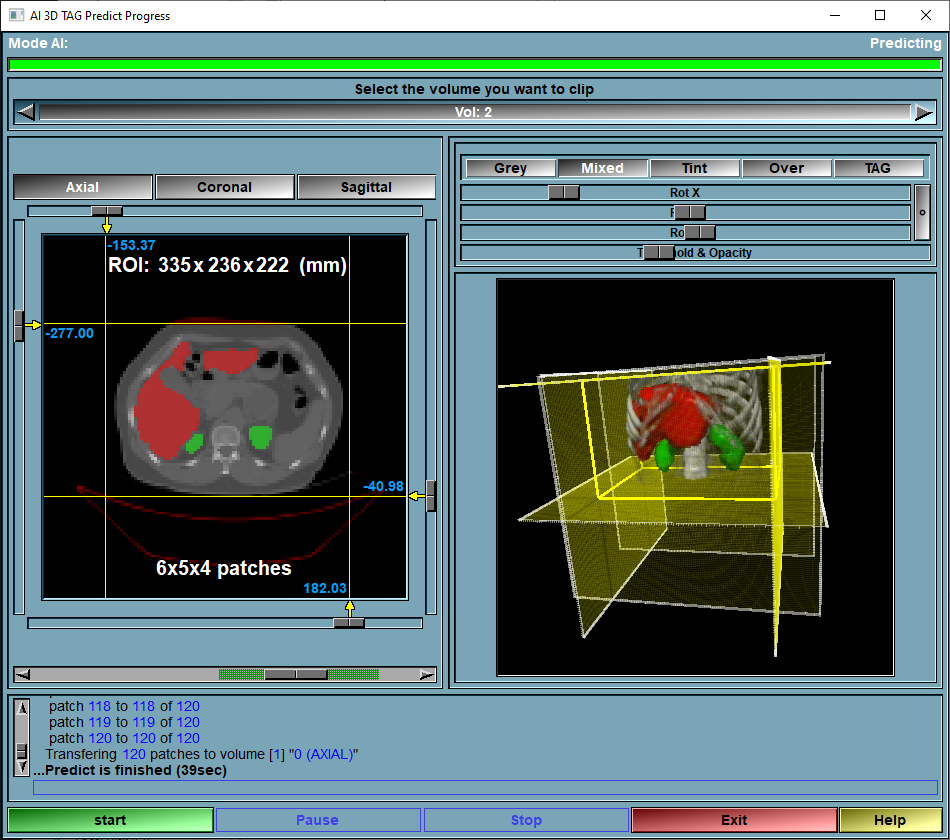
The Predict page
This is the page where the magic happens!
From the AI list, select the AI you want to use. Alternatively, you can use the Config page to select a model and its assciated weights.
Select the desired images in sliceOmatic and click on the "Compute" button.
Note:
|
|
|
|
|
|
AI Selection |
Select the desired AI from the list. This will automatically set the Model and the associated Weights.
|
Rename |
Rename the files associated with the selected AI (script, text and AI weights).
|
Delete |
Delete the files associated with the selected AI.
|
Config |
Open the Configuration window
|
Compute |
Open the "Predict Compute" Window.
|
The goal of the AI module is to compute the segmentation automatically. This is done here.
The prediction module is the one that compute the segmentation. The segmentation is computed from the selected weights chosen in the "Predict" tab (or from the "Config" menu). The training has been done with patches of a specific size and overlap. The loaded volumes will also be subdivided in patches and the prediction will be done on each sequentially.
You can limit the ROI of the volume that will be used for the prediction with the interface.
If the selected weights have been trained with multiple classes and you only want to get the prediction for one tissue, you can use the "TAG Lock" tool to protect the other tissues.
|
|
|
Progress Bar |
Show the progression of the predicting
|
Volume selection |
If multiple volumes are loaded in sliceOmatic, you can select which of these will be displayed and clipped in the volume display window
|
Volume display 2D |
Show the selected volume in 2D. From this window you can limit the volume that will be predicted.
|
Volume display 3D |
Give a 3D view of the selected volume with a view of the clip planes used to limit the ROI used for prediction.
|
Text Feedbacks |
Report some info on the progression of the prediction.
|
Start |
Start the computation of the prediction.
|
Pause/Resume |
Pause/resume the computation.
|
Stop |
Stop the computation.
|
Exit |
Exit this page.
|
Help |
Display this web page. |
The Python files
You can add your own metrics and models to this mode. You just have to follow the syntax described here:
The Python directory.
All the Python files used by sliceOmatic's AI module are stored in the Python directory. By default this is in "c:\Program Files\TomoVision\SliceO_Python". The location of this directory can be changed through the sliceOmatic's "Config" interface.
The Metrics Python files.
For my metrics files, I use names that start with "metrics ", but the name is not really important. What is important is that the file contain either a "metrics_2D" function, a "loss_2D" function or both.
You can have 3 types of metrics functions
The "Loss" metrics:
- The Loss functions compute a loss metrics used to train the AI.
The value of the loss function must decrease when we are approach perfect results.
The name of the loss function must be: "loss_3D_TAG" and have 2 arguments: "y_true" and "y_pred", 2 tensors, as described in Keras doc.
You must define a flag for the function: loss_3D_TAG.flag =... (The choices are between: "mean" "weighted" and "class" )
You must define the name of the function: loss_3D_TAG.__name__ =...
You must define a version for the function: loss_3D_TAG.__version__ =... (The version syntax is: "major.minor". ex: "2.3")
You must define the description of the function: loss_3D_TAG.description =...
|
|
import keras as kr
# ---------------------------------------------------- # The function must be called "loss_3D_TAG" # ---------------------------------------------------- def loss_3D_TAG( y_true, y_pred ) :
# --- We simply call the Keras function --- return( kr.losses.categorical_crossentropy( y_true, y_pred ) )
# ---------------------------------------------------- # Give a name for this metrics # ---------------------------------------------------- loss_3D_TAG.__name__ = "Categorical CrossEntropy"
# ---------------------------------------------------- # Give a version for this metrics # ---------------------------------------------------- loss_2D_TAG.__version__ = "2.1"
# ---------------------------------------------------- # Give a name for this metrics # ---------------------------------------------------- loss_3D_TAG.description = "Call the Keras \"categorical_crossentropy\" function:\n" \ "Computes the crossentropy loss between the labels and predictions." |
You can have 2 types of metrics: "global" or "Per class".
The "global" metrics:
- The Metrics functions compute a metrics used as feedback when training or testing.
The name of the metrics function must be: "metrics_3D_TAG" and have 2 arguments: "y_true" and "y_pred", 2 tensors, (as described in Keras doc.)
You must define a flag for the function: metrics_3D_TAG.flag =... (The choices are between: "mean" "weighted" and "class" )
You must define the name of the function: metrics_3D_TAG.__name__ =...
You must define a version for the function: metrics_3D_TAG.__version__ =... (The version syntax is: "major.minor". ex: "2.3")
You must define the description of the function: metrics_3D_TAG.description =...
|
|
|
The "per class" metric:
The name of the metrics function must be: "metrics_3D_TAG" and have 2 arguments: "class_idx" and "name", the index and name of the target class, for "Per class" metrics.
The "per class" metrics return a "global" metric function for the desired class.
You must define the name of the function: metrics_3D_TAG.__name__ =...
You must define a version for the function: metrics_3D_TAG.__version__ =... (The version syntax is: "major.minor". ex: "2.3")
You must define the description of the function: metrics_3D_TAG.description =...
You must also define a "flag" that must contain the string "class". (metrics_3D_TAG.flag = "class")
|
|
|
The Model Python files.
Again, I use the prefix "model " for my models files but the name is no important. The important is that the file contain a "model_3D_TAG" function.
That function receive 4 arguments: dim_x, dim_y, dim_z and num_classes. dim_x and y are the X and Y resolution of the images we will train this model with and num_classes is the number of classes. The function must define the layers of your model and return the "model" variable returned by the Keras "Model" function.
You must define the name of the function: model_3D_TAG.__name__ =...
You must define a version for the function: model_3D_TAG.__version__ =... (The version syntax is: "major.minor". ex: "2.3")
You must define the description of the function: model_3D_TAG.description =...
|
|
|
The Normalization and Dropout parameters
When using the Normalization parameter in your models, you must first access it from the sliceOmatic system variable: "$AI_NORMALIZATION". This integer value will be either 0:no normalization, 1:batch, 2:group or 3:instance.
The dropout is a float value accessed from the "AI_DROPOUT" sliceOmatic variable.
Here's is the suggested syntax:
|
|
|
Note:
|
|
|
The Test Python files.
By default, I test for the presence of Python, Numpy, Tensorflow and Keras. If you use another module in your Python code, you can add a new test file. SliceO will use this to test that the desired module is indeed loaded when it start the AI module.
The file must contain a "version" function that return the version number for the target module.
It must also contain a "__name__" and a "__version__" definition.
|
|
|
Accessing sliceOmatic variables from Python.
You can access any of the sliceOmatic variables from your python code.
The functions for this are:
sliceO_Var_Get_Int( Variable_Name )
sliceO_Var_Get_Float( Variable_Name )
sliceO_Var_Get_Str( Variable_Name )
return one (or an array of) value(s), depending on the variable.
sliceO_Var_Set_Int( Variable_Name, Value )
sliceO_Var_Set_Float( Variable_Name, Value )
sliceO_Var_Set_Str( Variable_Name, Value )
Assign the value "Value" to the variable "Var_Name". At this time only variable with one value can be "Set" (no arrays).
From the Display Area
There is no display area interaction specific to this mode.
From the Keyboard
The following commands are mapped to keyboard keys as a shortcut:
|
|
|
|
|
|
Key map |
Action |
|
|
|
|
|
|
F1 |
Set the color scheme mode for all windows: F1=Grey, F2=Mixed, F3=Tint and F4=Over |
|
|
F2 |
|
|
|
F3 |
|
|
|
F4 |
|
|
|
|
|
|
|
|
From the Command Line
System Variables defined in this library:
|
|
$AI_RES_X |
(I16) |
|
|
|
$AI_RES_Y |
(I16) |
|
|
|
$AI_RES_Z |
(I16) |
|
|
|
$AI_CLASS_NB |
(I16) |
|
|
|
$AI_CLASS_LABEL |
A(W) |
* $AI_CLASS_NB |
|
|
$AI_CLASS_RATIO |
A(F32) |
* $AI_CLASS_NB |
|
|
$AI_CLASS_WEIGHTS |
A(F32) |
* $AI_CLASS_NB |
Tversky metrics parameters:
|
|
$AI_TVERSKY_ALPHA |
(F32) |
|
|
|
$AI_TVERSKY_BETA |
(F32) |
|
Normalization and Dropout parameters:
|
|
$AI_NORMALIZATION |
(U8) |
|
|
|
$AIDROPOUT |
(F32) |
|
TAG re-mapping:
|
|
$AI_TAG_MAP_IN_NB |
(I32) |
|
|
|
$AI_TAG_MAP_IN_VAL |
A(U8) |
* $AI_CLASS_NB |
|
|
$AI_TAG_MAP_OUT |
A(U8) |
* $AI_CLASS_NB |
Training Parameters:
|
|
$AI_TRAIN_STEP_MAX |
(I32) |
|
|
|
$AI_TRAIN_STEP_CUR |
(I32) |
|
|
|
$AI_TRAIN_EPOCH_MAX |
(I32) |
|
|
|
$AI_TRAIN_EPOCH_CUR |
(I32) |
|
|
|
$AI_TRAIN_BATCH_MAX |
(I32) |
|
|
|
$AI_TRAIN_BATCH_CUR |
(I32) |
|
|
|
$AI_TRAIN_LR |
(F32) |
|
|
|
$AI_TRAIN_EARLY_FLAG |
(I32) |
|
|
|
$AI_TRAIN_EARLY_NB |
(I32) |
|
Python Variables:
|
|
$PYTHON_FLAG |
(U32) |
|
|
|
$PYTHON_MODULE_PATH |
(W) |
|
|
|
$PYTHON_INTERPRETER_PATH |
(W) |
|
|
|
$PYTHON_SLICEO |
(P) |
|
|
|
$PYTHON_TEMP |
(W) |
|
Commands recognized in this mode:
Python: path "path"
Python: temp "path"
A3T: file read "path"
A3T: file "path" (on|off)
A3T: preproc resolution x [y]
A3T: preproc modality value
A3T: preproc min value
A3T: preproc max value
A3T: preproc norm ("HU" | "min-max" | "z-score")
A3T: class nb value
A3T: class in id value
A3T: class out id value
A3T: class label id "label"
A3T: augment "name" (on|off) value
A3T: metrics loss "name"
A3T: metrics feedback clear
A3T: metrics feedback "name"
A3T: split test value
A3T: split mode value
A3T: split split value
A3T: split kfold value
A3T: model cur "name"
A3T: model modality value
A3T: model class value
A3T: weights cur "name"
A3T: weights load